Predicting gene expression levels from genomic sequences
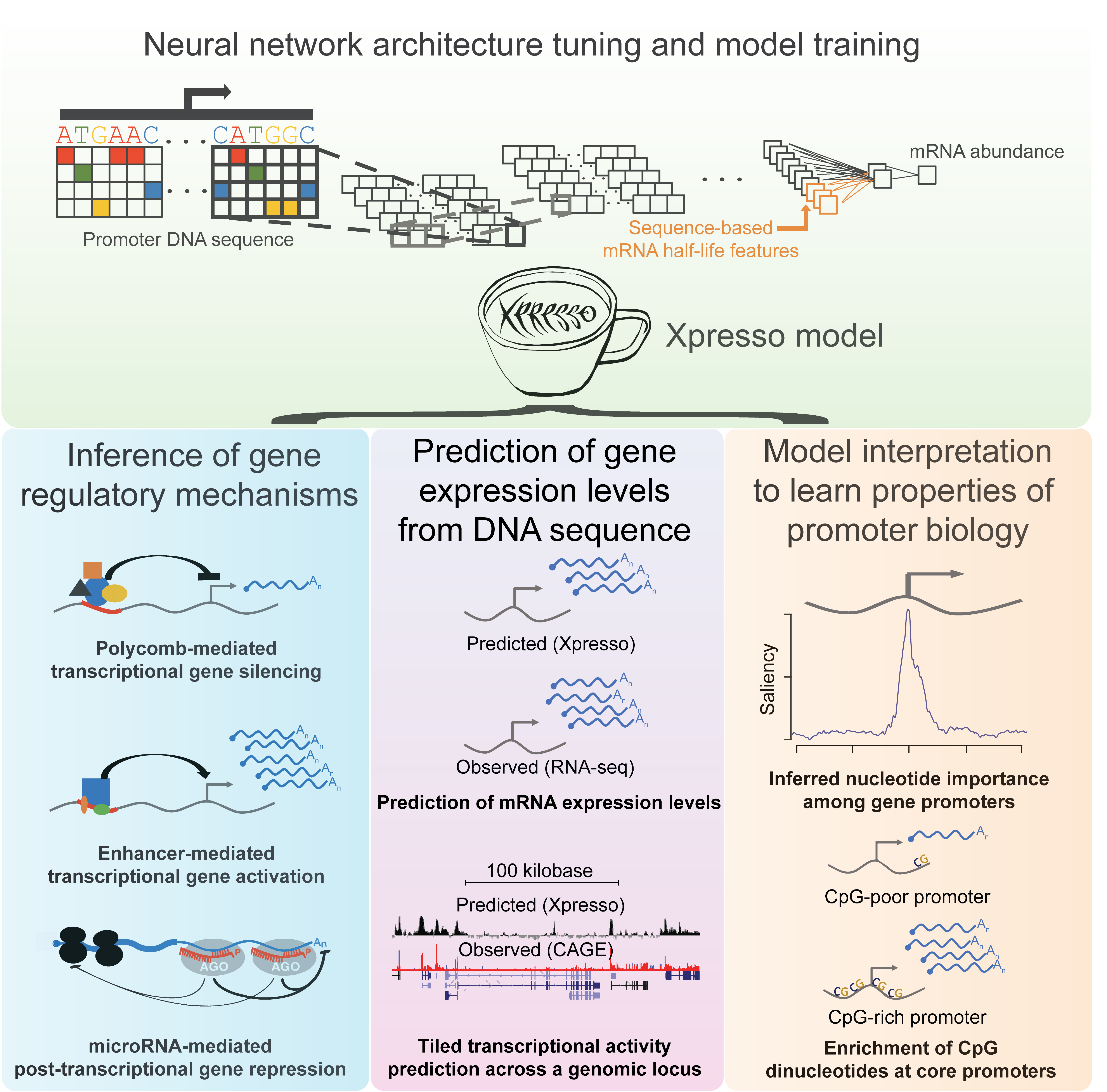
In our manuscript (Agarwal & Shendure 2020), we develop a suite of tools to predict gene expression levels from promoter DNA sequences and features related to mRNA half-life.

Xpresso relies upon training deep convolutional neural networks, which can be used to learn how the spatial relationships of motifs within DNA sequences predict gene expression levels.
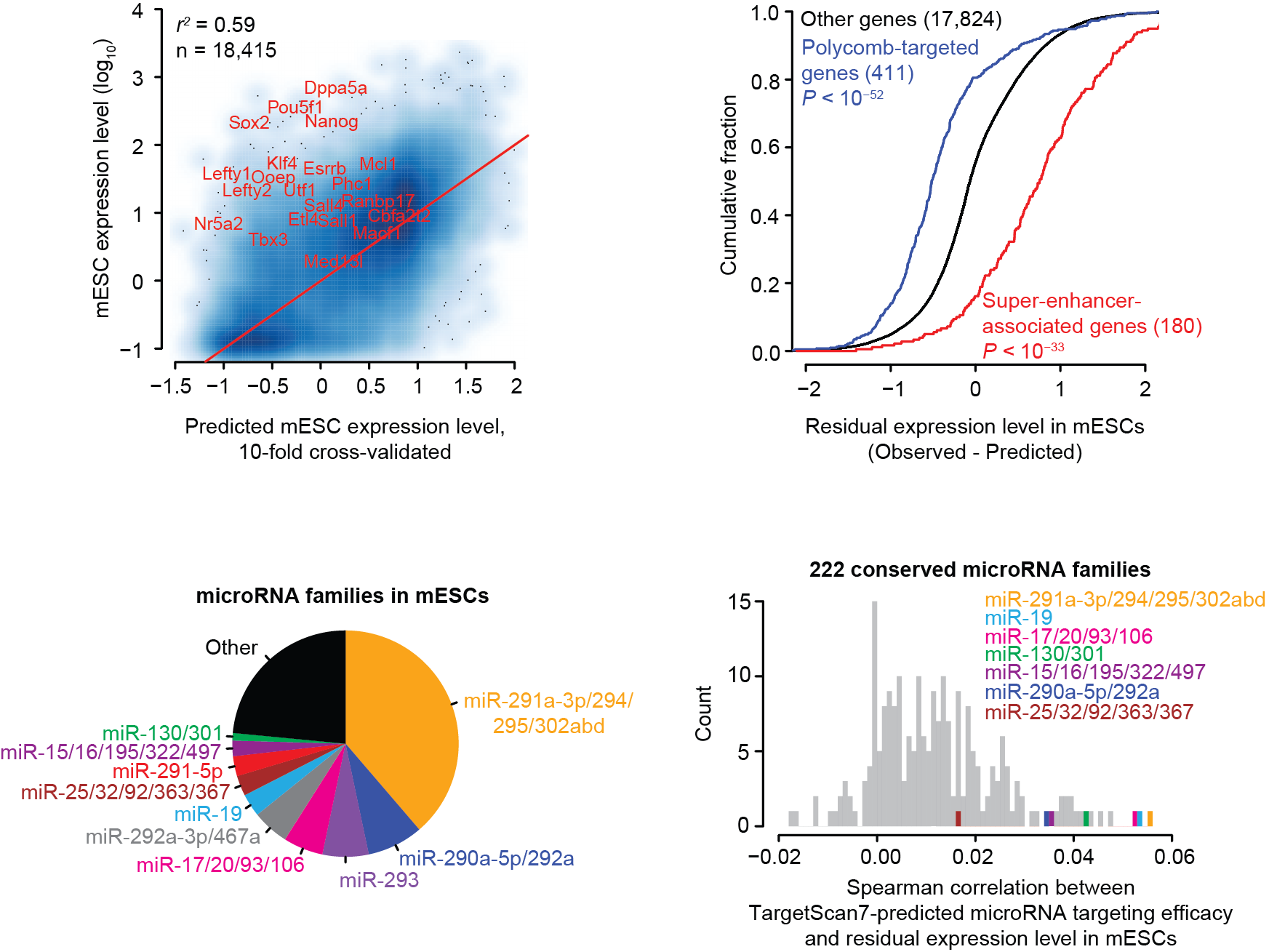
The residuals from our predictions can help generate hypotheses about gene regulatory mechanisms operating in cell types. For example, predictions in mouse ESCs reveal a number of super-enhancer-associated (SE) genes expressed more highly than predicted (top-left). Polycomb-targeted genes are repressed and SE genes are activated compared to our predictions (top-right). An analysis of TargetScan7-predicted microRNA targets implicates several conserved microRNA families as active (bottom-right) that match the most highly expressed microRNA families observed in mESCs (bottom-left).
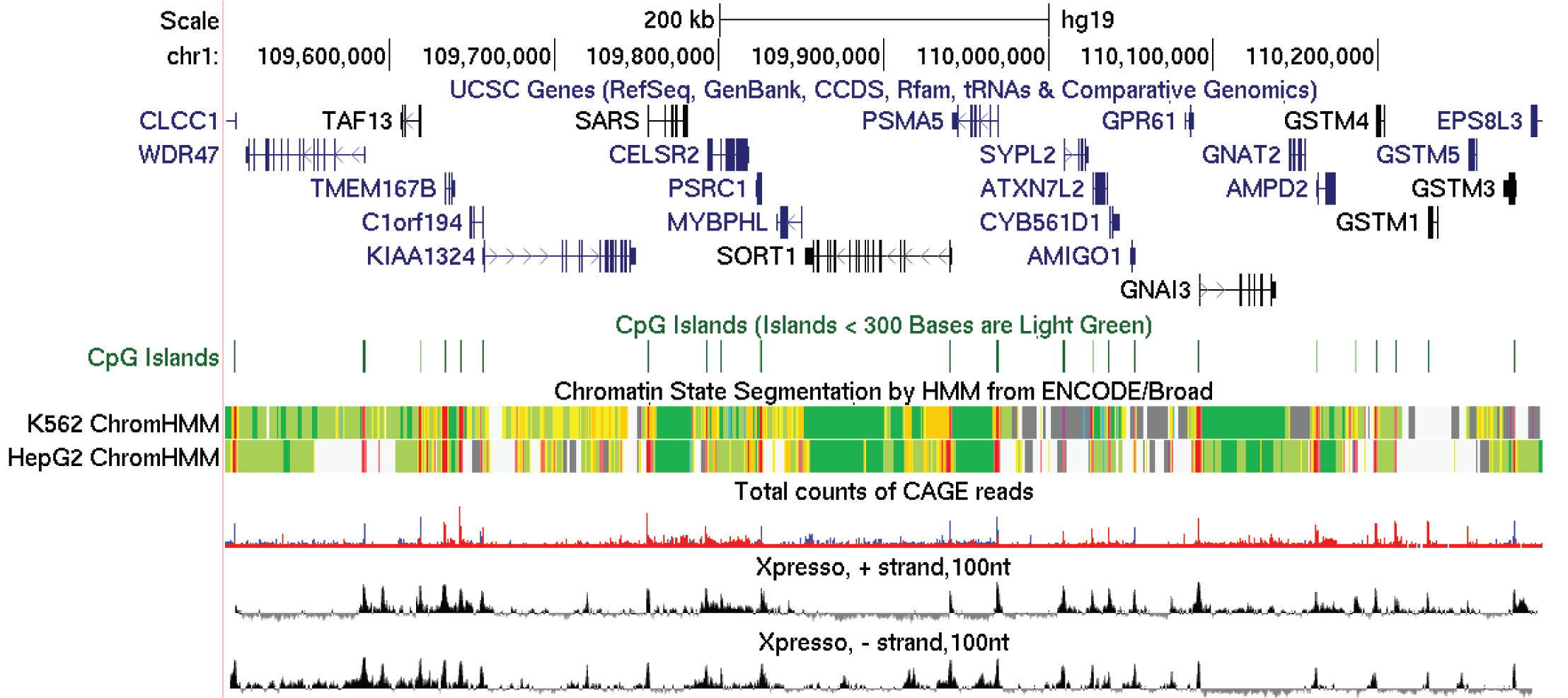
Alongside gene expression levels, Xpresso can predict transcriptional activity on arbitrary DNA sequences. The result above shows how prediction at 100nt steps across a genomic locus mirrors CAGE activity.
Downloads
We have pre-computed predictions for all human and mouse protein-coding genes, using models trained on among cell types in both species. Alternatively, you can download our code to generate your own predictions on arbitrary genomic regions or sequences with our pre-trained models.
Predictions on a variety of genomes and sequences
To learn how to use our code to generate predictions on your own sequences or genomic positions of interest for arbitrary mammalian species and cell types, please read this tutorial.
Train your own models
Finally, you can reproduce our results or train your own deep learning models on arbitrary cell types or species using our Github release or GPU-enable Colab.
Citation
If you find our work useful, please cite the following publication:
Agarwal V, Shendure J. Predicting mRNA abundance directly from genomic sequence using deep convolutional neural networks. 2020. Cell Reports 31 (7), 107663.
